GSP4PDB (Graph-based Structural Patterns for PDB) is a bioinformatics web tool that lets the users design, search and analyze protein-ligand structural patterns inside the Protein Data Bank (PDB).
GSP4PDB is formed by three main elements: gsp4pdb-extractor, a java tool which allows to extract and pre-process data from PDB files; a PostgreSQL database system which is used to store and manage the protein data used by the application; and a web application which provides a graphical interface for designing and querying graph-based structural graph patterns.
The novel feature of GSP4PDB is that a protein-ligand structural pattern is graphically designed as a graph such that the nodes represent protein’s components and the edges represent structural relationships. The resulting graph pattern is transformed into a SQL query, and executed in the PostgreSQL database system where the PDB data is stored. The results of the search are presented using a textual representation, and the corresponding binding-sites can be visualized using a JSmol interface.
GSP4PDB is available at https://structuralbio.utalca.cl/gsp4pdb/
A conference paper describing GSP4PDB is available here.
Graph-based structural patterns
A graph-based structural pattern is a graph where the nodes represent protein’s components (i.e. amino acids and ligands) and the edges represent structural relationships (distance between two amino acids, distance between a ligand and an amino acid, precedence relationship between two amino acids).
The following Figure shows an example of a simple graph-based structural pattern representing a protein-ligand interaction.
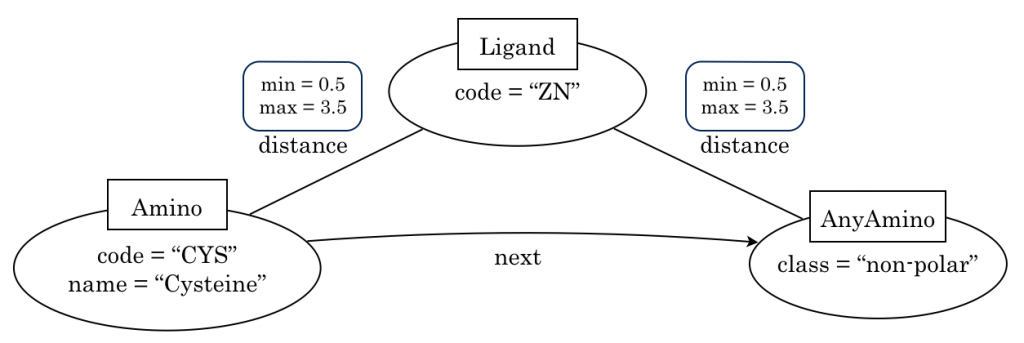
The above Figure shows the three types of nodes (Amino, AnyAmino and Ligand), the two types of edges (distance and next), and some properties for them.
Graphical- interface for protein-ligand interactions
GSP4PDB includes a web-based graphical interface which allows to design protein-ligand interactions as graphs. The following Figure present the interface. On the left side is the design interface where the protein-ligand interaction can be “drawn”. On the right sise is the output interface where the “matched” binding sites are shown in textual form.
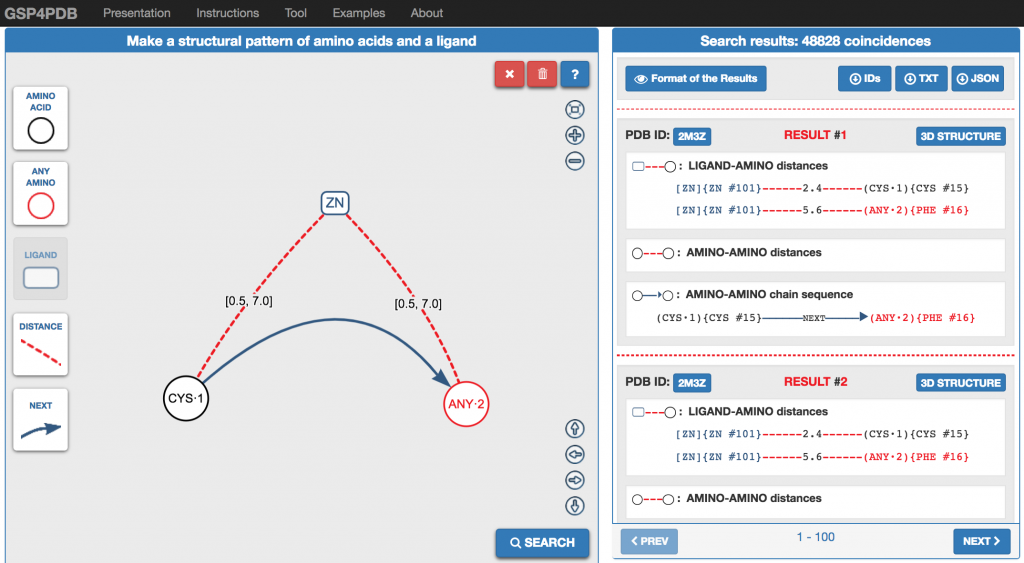
Protein Data Extraction and Pre-processing
GSP4PDB was thought to work using data obtained from the Protein Data Bank (PDB). In this sense, we have developed gsp4pdb-extractor, a command-line java application which allows to process PDB files, and export the protein data to the PostgreSQL database.
The single parameter of gsp4pdb-extractor is the directory where the PDB files are stored. The current version is restricted to process files encoded using the PDB format (*.pdb, *.ent or *.ent.gz). For each protein file, gsp4pdb-extractor parses the file using biojava4 and create an object model of the protein. The main classes of the model are Protein, SChain, Aminoacid, AminoStandard, AminoStandardList, Hetam (Ligand), AtomAmino and AtomHet.
During the creation of the object model, three distance measures (expressed in Angstroms) are pre-computed: DistanceAminoAmino, which is calculated as the distance between the alpha carbon atoms of two amino acids; and DistanceAminoHet, which corresponds to the distance between the alpha carbon atom of the amino acid and the center of mass of the ligand. Distances greater than 7.0 Amstrongs are not considered. In addition to the distance relationships, we define the class NextAminoAmino to represent the sort between each pair of amino acids in the chain.
After the object model of the protein is constructed, gsp4pdb-extractor loads the data to the PostgreSQL database by using bulks of 1000 SQL instructions.
Relational Schema used by GSP4PDB
GSP4PDB uses a PostgreSQL database system for storing and managing the protein data. The relational schema is given by the following tables:
- Protein (id, title, classification, organism, dep_date, technique, mod_date)
- Chain (id, protein_id, seqres, num_het, num_amino)
- aminoacid (id, chain_id, symbol, protein_id, next_amino)
- het (id, chain_id, symbol, protein_id, num_atom)
- distance_amino_amino (amino1_id, amino1_symbol, amino1_class, amino2_id, amino2_symbol, amino2_class, min, max)
- distance_het_amino (het_id, het_symbol, amino_id, amino_symbol, amino_class, min, max)
- next_amino_amino (amino1_id, amino1_symbol, amino1_class, amino2_id, amino2_symbol, amino2_class)
In practice, just the tables distance amino amino, distance het amino and next amino amino are necessary to search graph-based structural patterns. In fact, these tables contain data from other tables, hence, they introduce data redundancy. This unnormalized design is useful to improve query computation.
Note. We use Het instead of Ligand in order to maintain compatibility with the terms used by PDB.
Transforming graph-based structural patterns into SQL queries
The most challenging issue concerning the implementation of GSP4PDB was to define a method to transform a graph-based structural pattern into a SQL query. In general terms, we define a method which generates a SQL query expression for each node-edge-node structure in the graph pattern. The final SQL query, expressing the complete graph pattern, is the compositions of all the sub-expressions.
Next we present the transformations for every node-edge-node structure. In each SQL expression presented below, we will use <<parameter>> to denote a variable which will be replaced for an specific value, depending on the real graph pattern.
Ligand (Node1) – Distance (range) – Amino (Node2)
SELECT het_id, amino_id AS amino<<Node2_Id>>_id, amino_symbol AS amino<<Node2_Id>>_symbol, min AS min_het_amino<<Node2_Id>> FROM distance_het_amino WHERE het_symbol = "<<Node1_Symbol>>" AND amino_symbol = "<<Node2_Symbol>>" AND ((min < <<dmin>> AND max >= <<dmin>>) OR (min <= <<dmax>> AND max > <<dmax>>) OR (min >= <<dmin>> AND max <= <<dmax>>) OR (min < <<dmin>> AND max > <<dmax>>))
Ligand (Node1) – Distance (range) – Any (Node2)
SELECT het_id, amino_id AS amino<Node1_Id>>_id, amino_symbol AS amino<<Node1_Id>>_symbol, min AS min_het_amino<<Node1_Id>> FROM distance_het_amino WHERE het_symbol = "<<Node1_Code>>" AND amino_class = "<<Node2_Class>>" AND ((min < <<dmin>> AND max >= <<dmin>>) OR (min <= <<dmax>> AND max > <<dmax>>) OR (min >= <<dmin>> AND max <= <<dmax>>) OR (min < <<dmin>> AND max > <<dmax>>))
Amino (Node1) – Distance (range) – Amino (Node2)
SELECT * FROM (SELECT amino1_id AS amino<<Node1_Id>>_id, amino1_symbol AS amino<<Node1_Id>>_symbol, amino2_id AS amino<<Node2_Id>>_id, amino2_symbol AS amino<<Node2_Id>>_symbol, min AS min_amino<<Node1_Id>>_amino<<Node2_Id>> FROM distance_amino_amino WHERE ((min < <<dmin>> AND max >= <<dmin>>) OR (min <= <<dmax>> AND max > <<dmax>>) OR (min >= <<dmin>> AND max <= <<dmax>>) OR (min < <<dmin>> AND max > <<dmax>>)) AND amino1_symbol = "<<Node1_Code>>" AND amino2_symbol = "<<Node2_Code>>" UNION SELECT amino2_id AS amino<<Node1_Id>>_id, amino2_symbol AS amino<<Node1_Id>>_symbol, amino1_id AS amino<<Node2_Id>>_id, amino1_symbol AS amino<<Node2_Id>>_symbol, min AS min_amino<<Node1_Id>>_amino<<Node2_Id>> FROM distance_amino_amino WHERE ((min < <<dmin>> AND max >= <<dmin>>) OR (min <= <<dmax>> AND max > <<dmax>>) OR (min >= <<dmin>> AND max <= <<dmax>>) OR (min < <<dmin>> AND max > <<dmax>>)) AND amino1_symbol = "<<Node1_Code>>" AND amino2_symbol = "<<Node2_Code>>" ) AS q_<<Node1_Id>>_<<Node2_Id>>
Amino (Node1) – Distance (range) – Any (Node2)
SELECT *
FROM
( SELECT
amino1_id AS amino<<Node1_Id>>_id,
amino1_symbol AS amino<<Node1_Id>>_symbol,
amino2_id AS amino<<Node2_Id>>_id,
amino2_symbol AS amino<<Node2_Id>>_symbol,
min AS min_amino<<Node1_Id>>_amino<<Node2_Id>>
FROM distance_amino_amino
WHERE
((min < <<dmin>> AND max >= <<dmin>>) OR
(min <= <<dmax>> AND max > <<dmax>>) OR
(min >= <<dmin>> AND max <= <<dmax>>) OR
(min < <<dmin>> AND max > <<dmax>>)) AND
amino1_symbol = "<<Node1_Code>>" AND
amino2_class = "<<Node2_Class>>"
UNION
SELECT
amino2_id AS amino<<Node2_Id>>_id,
amino2_symbol AS amino<<Node2_Id>>_symbol,
amino1_id AS amino<<Node1_Id>>_id,
amino1_symbol AS amino<<Node1_Id>>_symbol,
min AS min_amino<<Node1_Id>>_amino<<Node2_Id>>
FROM distance_amino_amino
WHERE
((min < <<dmin>> AND max >= <<dmin>>) OR
(min <= <<dmax>> AND max > <<dmax>>) OR
(min >= <<dmin>> AND max <= <<dmax>>) OR
(min < <<dmin>> AND max > <<dmax>>)) AND
amino2_symbol = "<<Node1_Code>>" AND
amino1_class = "<<Node1_Class>>"
)
AS q_<<Node1_Id>>_<<Node2_Id>>
Any (Node1) – Distance (range) – Any (Node2)
SELECT *
FROM
( SELECT
amino1_id AS amino<<Node1_Id>>_id,
amino1_symbol AS amino<<Node1_Id>>_symbol,
amino2_id AS amino<<Node2_Id>>_id,
amino2_symbol AS amino<<Node_Id>>_symbol,
min AS min_amino<<Node1_Id>>_amino<<Node2_Id>>
FROM distance_amino_amino
WHERE
((min < <<dmin>> AND max >= <<dmin>>) OR
(min <= <<dmax>> AND max > <<dmax>>) OR
(min >= <<dmin>> AND max <= <<dmax>>) OR
(min < <<dmin>> AND max > <<dmax>>)) AND
amino1_class = "<<Node1_Class>>" AND
amino2_class = "<<Node2_Class>>"
UNION
SELECT
amino2_id AS amino<<Node1_Id>>_id,
amino2_symbol AS amino<<Node1_Id>>_symbol,
amino1_id AS amino<<Node2_Id>>_id,
amino1_symbol AS amino<<Node2_Id>>_symbol,
min AS min_amino<<Node1_Id>>_amino<<Node2_Id>>
FROM distance_amino_amino
WHERE
((min < <<dmin>> AND max >= <<dmin>>) OR
(min <= <<dmax>> AND max > <<dmax>>) OR
(min >= <<dmin>> AND max <= <<dmax>>) OR
(min < <<dmin>> AND max > <<dmax>>)) AND
amino1_class = "<<Node2_Class>>" AND
amino2_class = "<<Node1_Class>>"
)
AS q_<<Node1_Id>>_<<Node2_Id>>
Amino (Node1) – Next – Amino (Node2)
SELECT amino1_id AS amino<<Node1_Id>>_id, amino1_symbol AS amino<<Node_Id>>_symbol, amino2_id AS amino<<Node2_Id>>_id, amino2_symbol AS amino<<Node2_Id>>_symbol FROM next_amino_amino WHERE amino1_symbol = "<<Node1_Code>>" AND amino2_symbol = "<<Node2_Code>>"
Amino (Node1) – Next – Any (Node2)
SELECT amino1_id AS amino<<Node1_Id>>_id, amino1_symbol AS amino<<Node1_Id>>_symbol, amino2_id AS amino<<Node2_Id>>_id, amino2_symbol AS amino<<Node2_Id>>_symbol FROM next_amino_amino WHERE amino1_symbol = "<<Node1_Code>>" AND amino2_class = "<<Node2_Class>>"
Any (Node1) – Next – Amino (Node2)
SELECT amino1_id AS amino<<Node1_Id>>_id, amino1_symbol AS amino<<Node1_Id>>_symbol, amino2_id AS amino<<Node2_Id>>_id, amino2_symbol AS amino<<Node2_Id>>_symbol FROM next_amino_amino WHERE amino2_symbol = "<<Node2_Code>>" AND amino1_class = "<<Node1_Class>>"
Any (Node1) – Next – Any (Node2)
SELECT amino1_id AS amino<<Node1_Id>>_id, amino1_symbol AS amino<<Node_Id>>_symbol, amino2_id AS amino<<Node2_Id>>_id, amino2_symbol AS amino<<Node2_Id>>_symbol FROM next_amino_amino WHERE amino1_class = "<<Node1_Class>>" AND amino1_class = "<<Node2_Class>>"